Overview |
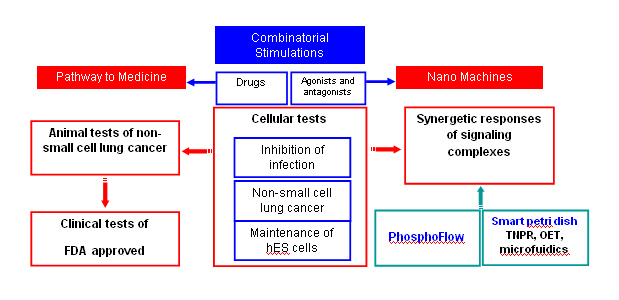
The goal of NDC is to characterize and quantify nano-molecular components of cells and to precisely control and manipulate these molecules and supramolecular assemblies to improve human health. The Center for Cell Control's (CCC) strategy to achieve this vision is to precisely control cell function with combinatorial drugs aimed at specific outcomes and measuring signaling complexes by PhosphoFlow or by Tunable Nan o Plasmonic Resonators (TNPR) in living cells to ultimately advance therapeutic goals. Our platform integrates real-time molecular detection with cell control. This approach enables response quantification of signaling nanomachineries, such as protein-protein interactions and signal transduction network activity, that guides cell fate under multiple types of stimulation. Using this integrative approach, synergistic or antagonistic interactions between molecular complexes are deciphered by quantifying multiple phosphorylation nodes in the signaling network. This strategy has already been successfully demonstrated and is beginning to link specific signaling nanomachines to translational medicine. Successful examples include in-vitro experiments to inhibit DNA virus infections, to eradicate cancer cells, and to maintain human embryonic stem (hES) cell pluripotency and self-renewal under defined culturing conditions.
|
NDC Goals |
NDC's overall goal is to:
• Characterize quantitatively molecular scale components or nanomachineries of the cell. This nanomedicine initiative is distinguished from other NDCs by its long-term focus on characterizing cellular processes and their interactions at a level of precision that has not been achieved to date.
• Precisely control and manipulate these molecules and supramolecular assemblies in living cells.
• Improve human health.
CCC has developed specific strategy to realize the NDC goal. Our plan is to:
• Manipulate relevant signal complexes by multiple stimulations for therapeutic purposes.
• Precisely measure system responses of signaling complexes by first using PhosphoFlow technology and then by tunable plasmonic resonators (TNPR) in living cells.
• Initiate the process of developing gene profile drug cocktail treatment on non-small cell lung cancer through the clinical collaboration project for improving human health.
|
Introduction |
Based on our strategy, CCC will realize its vision using the overall plan illustrated here. Combinatorial drug studies have shown potential for a rapid path to translational medicine and also provide a rich information data base for exploring the responses of signaling complexes, or signalosomes, in live cells. Over the past years, CCC has made two milestone advances:
• Identifying relevant signaling complexes in various settings and quantifying their responses, as measured by phosphorylation activity, with single or multiple drug stimulations. Thus, CCC has demonstrated specific coupling between “nano” and medicine.
• Removing the hurdle of tunable nanoplasmonic resonator (TNPR) mass production. This is an important step toward real-time measurements of signaling complex assembly/disassembly and network activity in live cells.
|
NDC Accomplishments |
I : Manipulating relevant signal complexes by multiple stimulations for therapeutic purposes
CCC's first task is to manipulate cell function with combinatorial drugs aimed at specific cellular outcomes or phenotypes, to ultimately advance therapeutic goals. It has been extremely challenging for clinical researchers to determine optimal doses of drugs to use when combined with each other. If a researcher were to test ten different concentrations of six drugs in every possible arrangement, he/she would have one million experimental conditions. We applied feedback system control technique and succeeded in removing this major road block. With only tens of iterative tests, we were able to identify optimized combinatorial drugs. In addition, the dosages of some drugs can be one order of magnitude lower than that of single drug usage. Currently we are working on in-vitro experiments to inhibit virus infections, eradicate cancer cells, and maintain human embryonic stem (hES) cell pluripotency and self-renewal under defined culturing conditions. The inhibition of viral infection VSV and the reactivation of KSHV have been published (ref. 13 and 29).
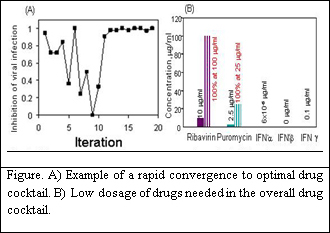
I-1: Optimizing Drug Combination for inhibiting vesicular stomatitis virus (VSV) infection
Viruses replicate by infecting a host cell and using the host cell's molecular machinery to create new progeny virions. Consequently, many of the molecular components of the virus and the host cell are similar. Therapeutics that target virus replication thus represent a double edged sword in that though they can efficaciously block viral replication they may also interfere with normal host cellular metabolism. Indeed, many anti-viral therapeutics that work effectively in vitro suffer in the clinical setting by toxic side effects. A central question in the effective clinical management of viral infection thus remains: is there an optimal method to treat a patient by inhibiting viral replication and still limit or abolish the toxic side effects of the drugs. To limit the side effects of high dose anti-viral therapies, treatments have entailed combinatorial drug use where multiple anti-viral drugs work in concert to limit the replication and spread of virus. We used 5 drugs with 10 concentrations to inhibit SVS infection of NIH 3T3 fibroblast cells. Three of these drugs were interferons (IFN), including IFNa, IFNb, and IFNg. IFNs are naturally produced proteins that are highly effective in organisms to block virus infections by producing an antiviral state. In addition, we used Ribavirin, a broad spectrum anti-viral drugs that is well characterized to stop RNA viruses like VSV. Also, we used puromycin a nonselective terminator of protein synthesis that at low doses can stop the rapid growth of viruses. The property of rapid convergence of the Gur Game is invaluable for regulating complex biological systems. Inhibition of the viral activity, which is defined as the percentage of cells not expressing GFP after incubation with VSV for 13 hours, was considered as the systemic outputs for the optimization of the antiviral activity in the host cells. In the experiment, three sets independent tests were carried out and all rapidly converged to the potent combination again in tens of iterations. Most dramatically, this approach has defined a potent drug combination using a combination of drugs at concentrations ten times less than individual drugs. This will definitely lead to an overall goal of creating a drug combination that is far less toxic while still as effective as high dose combinations.
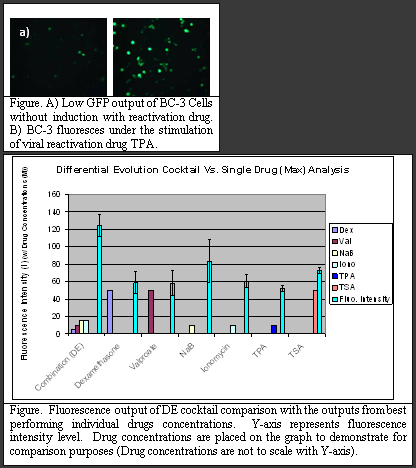
I-2: Optimizing Drug Combination for KSHV Reactivation
Understanding and controlling the reactivation of viruses are important for developing new viral therapies. In this study, we aimed to maximize the reactivation of Herpesvirus 8 or Kaposi's Sarcoma-associated Herpesvirus (KSHV) in a GFP transfected body cavity-based lymphoma cell line (BC-3) (Figure). A cocktail with 6 reactivation drugs and 10 concentrations requires 1 million tests which is extremely laborious and time consuming. By using the feedback control scheme, we have demonstrated in several different biological systems that twenty to thirty iterations is sufficient to finding the optimal drug cocktail within millions of possible combinations. At the heart of the closed-loop feedback control scheme is the mathematically well defined stochastic search algorithm that analyzes the system information and establishes the most effective driving inputs, i.e. drug combination.
To implement this KSHV reactivation study, a microfluidic platform was developed that has the capability of live cell imaging and precise cellular environment and stimulation control. Two search algorithms, Gur game and Differential Evolution, were used for the feedback control scheme. In this study, the feedback scheme was able to locate the optimal combination of drugs to reactivate KSHV with less than 30 test iterations for either of the two algorithms. The optimal cocktail was able to perform 49% better than the best performing single drug. Furthermore, the dosage needed in the drug cocktail is much lower that used in single drug cases.
II : Precisely measure system responses of signaling complexes
The simultaneous stimulations of relevant signaling nanomachines, such as protein-protein interactions and signal transduction network activities, have been successfully demonstrated to guide cell fate. We are integrating real-time molecular detection, Phosphoflow and TNPR, with cell control. Using this integrative approach, synergistic or antagonistic interactions between molecular complexes are deciphered by quantifying multiple phosphorylation nodes in the signaling network. This strategy has already been successfully demonstrated and is beginning to link specific signaling nanomachines to translational medicine.
II-1: PhosphoFlow Techniques for Quantifying Signaling Nanomachines
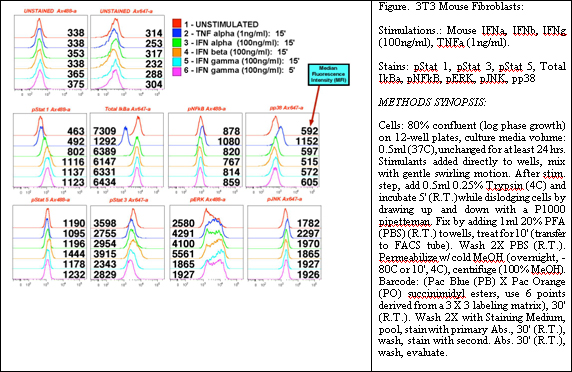
Phosphorylation of proteins plays an important role in cellular regulatory mechanisms. We have developed two complementary techniques to quantify the phosphorylation state of signaling protein complexes. In signaling biology, it has become clear that individual signaling molecules are not reflective of the state of a system. Multiple feedback mechanisms, redundancies, and cyclic events make modeling of cell reactivity to environmental influences, such as drugs, particularly daunting. Moreover, the state of the signaling machinery at any given point in time is both a reflection of its history as well as a current position to respond depending upon the environmental stimulus. Systems biologists and network specialists in biomedicine have begun to define signaling complexes, or at least begin identifying the elements of pathways that act as surrogates for the state of a particular pathway. This has led to approaches that both enhance our representations of signaling networks, as well to developing tools that use network states as 'targets' for drug action. Clearly, since viruses and cancer work at the level of modulating the network state, we as biomedical engineers (in the sense that we are attempting to modulate such network states back to health) must understand the means by which networks regulate themselves.
Given the nature of such complex systems, the evolution of eukaryotes in particular has chosen a path of supramolecular complexes with transient interchangeable parts as the basic units of decision-making within cells. Some protein elements of such machine complexes remain as core features of these nanomachines, while others are transiently associated with the nanomachines as they alter the basic function of the nanomachines, or are altered themselves. To faithfully reflect the biology and mechanism of such supramolecular machines and the networks in which they operate, we have chosen to quantify the functional states of many of the sub-elements of these systems via PhosphoFlow techniques.
For instance, NF- k B remains associated with I k B until one of its components (I k B) is phosphorylated. This leads to a dissociation of the NF- k B heterodimer and subsequent phosphorylation of the p65 subunit of that complex. Prior to activation, another supramolecular complex of kinases and inhibitors play a similar and more integrative role by interacting with numerous cellular pathways that impact NF- k B activity. Other examples abound, including the scaffolding proteins of the MAP Kinase pathways (STE5, KSR for MAP-RAS interactions, and MP1) and STAT dimer associations on cytoplasmic tails that lead to their activation and complexes that control the cell cycle. All of these systems must be described not just as the functions of their individual components, but at a level that faithfully reflects the mutable nature of their interactions and the nanomachinery to which they contribute and affect.
We have established approaches and protocols for measuring individual cell phosphorylation and correlations of such events to network states and phenotypic outcomes in response to drug treatments. Coupling of these developed approaches to known 'physical' interaction maps, scaffolds, and other defined nanomachinery within the cells (supramolecular complexes as noted above), will provide a new vision of how to represent the every shifting components of cellular signaling, and will allow us to define the multiple critical inflection points (multi-drug screening against networks as a whole) where modulation of activity of the network as a whole can proceed successfully.
II-2: Mass Production of TNPRs
We have successfully adapted nanoimprint lithography (NIL) for the mass fabrication of TNPRs. Using a NIL process that is designed for metal nanostructures, we have achieved reliable high-yield TNPR arrays over large areas the size of standard microscope slides, or equivalent to 2-3x10 9 TNPRs at a time. This capability presents two distinct opportunities. First, large-area substrates will enable massive improvement of the throughput of TNPR enhanced extracellular sensing of signaling molecule secretion. This will be achieved by covering the TNPR substrates with arrays of microfluidic channels and studying numerous specimens on the same chip. Second, we can now produce TNPRs in quantities appropriate for ballistic injection (achieved with high viability in pilot studies) and intracellular investigations of signalosome activity. Our current fabrication focus is to continue improving the yield and quality of TNPR solubilization. And also we are continuing efforts in developing a biocompatible sacrificial film to release TNPRs from the substrate into an aqueous medium and the protocols of peptide functionlization for eventual intracellular delivery and signaling pathway sensing.

II-3: OET techniques for Membrane Perforation
The aim of this project is to develop an alternative technique for delivering TNPRs into the intracellular space to monitor phosphorylation events. We are pursuing two methods simultaneously that combine optoelectronic tweezers (OET) with poration capability.
The first technique involves electroporation. Electroporation is a common technique for the introduction of exogenous molecules across the, otherwise, impermeant cell membrane. Conventional techniques are limited by either low throughput or limited selectivity. We present a novel technique whereby we use patterned light to create virtual electrodes which can induce the parallel electroporation of single cells. This technique seamlessly integrates with OET to provide a single cell manipulation platform as well. Cells are placed in the OET device and are manipulated using patterned light. Then, through a change in device bias, only cells that are illuminated in the device are electroporated. We have successfully demonstrated the delivery of fluorescent molecules into the cell. Additionally, we have completed a study proving that cells remain viable following electroporation. Since we are able to perform this technique in parallel, with single cell resolution, we are able to extract out the optimal field dosage to deliver a molecule to a cell without harming it. These results have recently been published in the Royal Society of Chemistry's Lab on a Chip journal.
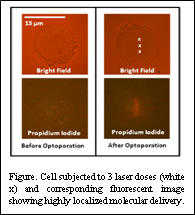
The second technique uses a femtosecond laser to create highly localized holes in the cell membrane. Due to short pulse widths, damage to the cell outside of the focal point of the laser beam is minimized. This allows us to selectively deliver molecules and particles to specific regions of the cell. We have successfully created pores ranging up to 500 nm in diameter in the cell membrane. We have used this technique to selectively deliver a fluorescent dye to only a small area of the cell. By combining this technique with the ability of OET to concentrate nanoparticles near a cell, we can create large concentrations of probes near the laser-induced membrane pore allowing for highly efficient and localized delivery. By combining OET with membrane poration capability, OET provides a means to efficiently deliver plasmonic probes (TNPRs) into the cell to monitor phosphorylation and other cellular kinetic events.
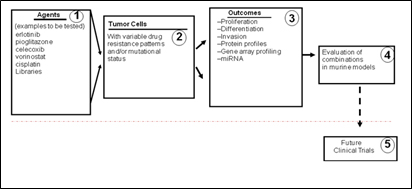
III: Drug cocktail treatment on non-small cell lung cancer through clinical collaboration
We are currently using techniques such as combinatorial drug cocktail technology and phosphor-flow technology to provide a pathway towards translational medicine. For lung cancer, we focus on assessing combination drug cocktail therapies tailored to gene array profiles, sensitivities of individual tumors, and lower toxicity to non-cancer lung cells. By using the data obtained from the Herpesvirus reactivation model we are also investigating the kinetics of cellular machines under combinational drug therapies. The integration of mathematic approaches for tailoring drug combinations, kinetic behavior of cellular response, and PhosphoFlow analysis techniques will allow a personalized medicine approach that targets susceptible pathways in individual patients and enable clinical translations of these investigations for patients with lung cancer.
III-1: Effect of five drug combinations on lung cancer cells and normal cells
We have determined the single drug dose response curve for (SAHA, Pioglitazone, Celecoxib, Cisplatinum and Erlotinib) selected out of the screened drugs (SAHA, Pioglitazone, Celecoxib, Cisplatinum, Erlotinib, Rapamycin, Paclitaxel, Lapatinib, Alimta, Erbitux and Velcade) on the lung cancer cell line A549 and the normal lung fibroblast cell AG02603. From the single drug dose response curves, we have selected five drugs (SAHA, Pioglitazone, Celecoxib, Cisplatinum and Erlotinib) and their concentration ranges to test their combinatory impact. SAHA is inhibitor of histone deacetylase (HDAC) 1 and 3; Pioglitazone selectively stimulates nuclear receptor peroxisone proliferator-activated receptor gamma (PPAR?); Celecoxib is a highly selective COX-2 inhibitor and primarily inhibits this isoform of cyclooxygenase (inhibition of prostaglandin production); Cisplatinum complexes are formed in cells, which bind and cause cross-linking of DNA that interferes with cell division -- ultimately triggering apoptosis, or programmed cell death; Erlotinib specifically targets the epidermal growth factor receptor (EGFR) tyrosine kinase. It binds in a reversible fashion to the ATP binding site of the receptor. These drugs have multiple cellular targets and their interactions are difficult to predict.
We have measured the cellular ATP levels in response to combinations of five drugs on A549 cells and the corresponding erlotinib-resistant A549T cells. To compare the drug effort on different kinds of cells, we also measured the cellular responses of a different lung cancer cell H358 cells and the normal lung fibroblast cell AG02603 (non-cancer cell as control). Mathematical models of the relationships between input drug combinations and output cellular responses were generated. Based on initial analysis we can see that there is a significant difference between the responses of normal and cancer cells to the same set of drugs combinations. Additionally, the drug-resistant cell line exhibited a response profile similar to the non-resistant cell line in response to treatment by combinations of drugs, which suggests that drug combinations can be effective in treating patients who have developed drug-resistance or to prevent the emergence of drug-resistance at the initiation of therapy. Once the results from the in vitro studies have been established we will move towards animal testing of the drug combination therapy.
|
Pathway to Medicine |
Combinatorial drug technology places the CCC at a strategic position for examining nanoscale activities, such as signaling complex phosphorylation, to rapidly move toward translational medicine. CCC received a clinical collaboration award headed by Dr. Steve Dubinett. This project is applying a rapid search scheme for lung cancer drug discovery that will move to small animal testing in the second year of effort.
Lung cancer is the leading cause of death in the United States and worldwide. It is estimated that lung cancer results in more than 1 million deaths per year worldwide and in the United States, causes approximately one death every 4 minutes. These devastating statistics suggest that an entirely new approach is necessary for the treatment and prevention of lung cancer. Although smoking cessation has made an impact in lung cancer incidence, former smokers remain at an elevated risk of lung cancer for many years. Our current research is to develop a novel system for the optimization of drug combinations for lung cancer therapy by utilizing an artificial neural network modeling algorithm. Our approach will determine optimal combinations of multiple drugs against panels of well characterized drug resistant lung cancer cell lines. These cell lines are being characterized both in terms of their gene array expression profiles and their responses to drugs so that we can ultimately define novel combination therapies targeted to specific genetic abnormalities, working through identified signalosomes.
|
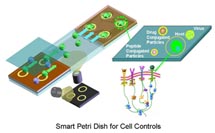
Smart Petri Dish with a Unique Suite of Nanomodalities |
Smart Petri Dish with a Unique Suite of Nanomodalities - Members of our Center have developed many unique nano/micro technologies which enable us to sense and manipulate individual molecule or cell for studying the cellular functions and for directing cells toward desired destiny. |
Nano/Micro Fluidics – backbone technology for bio-medical researches : Living subjects sustain their lives in fluid. The capability to move nano particles, bio-molecules or cells, of different species in a desired manner plays a major role in studying bio-medical sciences . The fluidic processes include moving, stopping and mixing of fluids as well as separating and concentrating embedded particles. Strong surface molecular effects and high viscous dissipation are the main challenges for handling fluids in micron-size bio-chemical reactors. Our group has extensively studied the rich nano/micro fluidic phenomena for 15 years, since the dawn of the microfluidics field. We examined the interactions of fluid and surface molecules. Various surface property controls have been developed for optimize the bio-chemical processing. Actuation schemes, including electrokinetic, hydrodynamic or magnetic forces, have been developed for controlling fluidic processes in micro/nano bio-reactors. ( http://ho.seas.ucla.edu/ ) |
 |
Optoelectronic tweezers (OET) - moving nanoparticles, cells and organelles with an optical micromachine : OET enables single-cell trapping at an operational power density that is one hundred thousand times less than that of a traditional laser tweezers setup. OET is based on a new mechanism called light-induced dielectrophoresis, which enables an optical beam to create a virtual electrode on a photoconductor-coated surface, producing a highly non-uniform electric field. The generation of a dielectrophoretic force locally by an optical beam allows for trapping, transporting, or sorting of cells with the potential to move particles or subcellular organelles down to 100 nm in size using optical power as low as 1 m W. ( http://www.eecs.berkeley.edu/%7Ewu/ ) |
 |
Nanoscope - seeing nanoscale events of live cells: For direct observations of molecular activities of live cells, a fundamental challenge to overcome is that the dimensions of biomolecules and their interactions are far below the diffraction limit of visible light (200 nm). Electron microscopes can probe nanoscale particles but not within living cells. We utilize materials with negative permittivity that amplify near-field optical waves with extremely short wavelengths. A “super-lens” based on this physical property can realize a “nanoscope” which has the potential to visualize the real-time motion of biomolecules, in their native state, at spatial resolutions far below the current limit. We are now able to spatially resolve images at 60 nm, such as microtubules, using a silver super-lens. The continued development of the nanoscope will enable direct visualization of objects at 10 nm resolution.( http://www.me.berkeley.edu/faculty/zhang/ ) |
 |
Sonocytology – feeling cell system responses: Several recent studies on the native oscillations and force required for membrane deformation by our members have established a new field termed “sonocytology”. An atomic force microscope (AFM) is used to probe the surface of cells under a variety of external and internal environmental conditions to obtain an oscillatory signal of nanomechanical origin. This oscillatory, periodic signal can be converted into sound and used as an indicator of cell health. The nanoscopic motion of the eukaryote yeast cell wall has been mechanistically linked to specific metabolic states, as well as responses to external perturbations and genetic differences. Sonocytology also enables cell damage detection; for example, when microtubule and actin dissociating agents used in chemotherapy are added, a change in cell elasticity is discernable much earlier than biochemical measurements of cell death. ( http://www.teitell-lab.com/ ) and ( http://www.chem.ucla.edu/dept/Faculty/gimzewski/ ) |
 |
Smart Petri Dish - integrated engineering system of nanomodalities for controlling biological cellular system : Cells and whole organisms detect, process, and respond to changes in their external and internal environments. Current methods for controlling and monitoring these responses in real time are largely lacking and form our main research motivation. In this center, we propose to use nanotechnology to characterize and manipulate intracellular functional molecules and to use engineering system principles to analyze and control regulatory circuitries of the cell. System control theories and feedback control principles that have been well established in the field of engineering can further offer insights and solutions in dissecting complex cellular network circuitries. |
 |
Home | People | Research | Collaboration | Communication | News | Jobs | Contact |